The long and the short of Amadeus’ investment in Oxford Nanopore Technologies
The Short
We are pleased to back Oxford Nanopore Technologies with a $24m secondary investment.
The Long
The journey leading to this investment started 20 years ago in a barn on the outskirts of Cambridge. At the time my partner Hermann Hauser asked me to help him carry out due diligence on a start-up called Solexa that had developed a novel technique combining: a new reversible terminator chemistry with laser technologies and advanced image processing that would revolutionise DNA sequencing. We were shown a demo by Solexa founders Shankar Balasubramanian and David Klenerman of their sequencing method based on fluorescently-labelled reversible terminators which enabled the identification of single nucleotides as they were washed over the DNA strands. After each cycle of terminator incorporation, lasers operating at different wavelengths were shone on the sample, enabling them to identify the individual nucleotides upon excitation and imaging of the specific fluorophores. The “eureka moment” they shared with us was captured in the image below – the equivalent of a pointillist’s rendition of life.
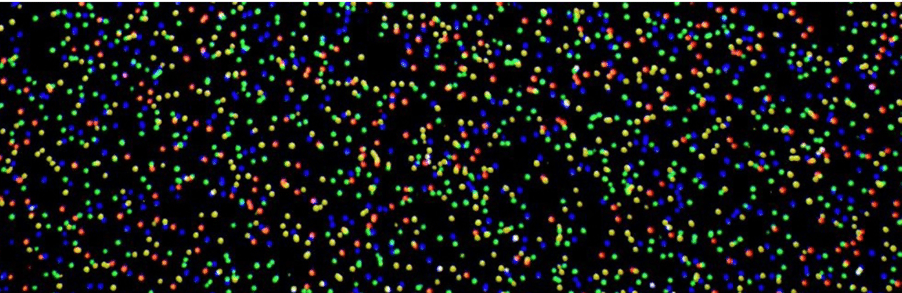
This picture represented the beginning of the current DNA sequencing market. We first invested in Solexa in 2001 and today that sector is worth close to $6B.
Solexa was eventually acquired by Illumina in 2007 for $650m. It became the core technology behind Illumina’s subsequent generations of sequencers; today’s the company’s market capitalisation is north of $62B, pointing to the potential of sequencing as a market.
Solexa’s technology eventually enabled the cost of DNA short-read sequencing to fall by a factor of close to 10,000. In short-read sequencing, data is generated by millions of short, individual DNA sequences (reads) ranging in length between 100 and 500 base pairs, with the majority at the lower end of the range. To put things in perspective, for a human genome sequence with good coverage 600 million reads of 150 bp are needed. For de novo sequencing, these short reads are assembled and analysed in order to re-construct the genome from which the reads originated. Given the scale of this endeavour, de novo assembly represents a very significant problem particularly when the reads are shorter and linking information between them is scant. Short sequences must therefore be mapped against unique positions in a reference genome to ensure fidelity of the assembly process. Reads often have sequencing errors, the reference sometimes has repeated elements and the orientation of a read relative to the reference is not known. Imagine putting together a multi-million pieces puzzle from a scrambled or low-resolution picture – now you get a sense of the challenge of short-read sequencing.
Despite these hurdles, and partly due to advancements in bioinformatics, over the last decade this technology has created a thriving eco-system of genetic data companies. Amadeus has been one of the most active investors in Europe in this field by backing leading companies like Healx, Congenica, Igenomix (recently sold to EQT) and more recently Veritas Int.
Meanwhile, we have been paying close attention to what Gordon Sanghera – founder and CEO of Oxford Nanopore Technologies – and team were trying to achieve. Outstanding talent was joining Oxford Nanopore, including some of the stars behind Solexa. Despite this, many industry observers were doubtful that it would be possible to overcome the enormous technical challenges of developing nanopore sequencing.
Oxford Nanopore’s sequencing technology is an electronics-based generation of sequencers that can be miniaturised and, in theory, sequence any length of DNA or RNA fragment, as well as potentially providing a platform to sense other molecules. As well as offering these novel features, such as the any-length reads, real-time data streaming and novel device formats, the technology would be expected to achieve similar performance to existing technologies in traditional metrics – cost and data quality. Over the past couple of years, it became increasingly obvious that Oxford Nanopore has been well on its way to achieve such performance with this new sequencing technology that we believe will significantly increase the size of the market; a much larger market that Oxford Nanopore has the potential to lead. Because of this we welcomed the opportunity to invest in the company as it scales.
Nanopore sequencing can process any length of DNA/RNA fragment, with a record to date of more than 4 million bases. Users choose to fragment their initial DNA sample to whatever length suits them. For some applications in human and plant genetics, scientists often want to sequence ultra-long fragments. Even if only choosing fragments around 50,000 bases long, this is still hundreds of times longer than traditional short reads, allowing them to characterise more variants and to assemble and phase the genome more easily. A recent study from deCODE in Iceland showed that when sequencing 3,622 human genomes with Oxford Nanopore’s PromethION, more than 20,000 structural variants were identified per genome, compared with 2-8,000 found using traditional SBS sequencing.
Oxford Nanopore’s unique technology works by monitoring changes to an ionic current as nucleic acids (i.e., nucleotides) are passed through a proprietary protein nanopore. The resulting signal is decoded to provide the specific DNA or RNA sequence -biology’s unique electrical signatures.

The technology platform is protected by more than 1,500 patents and patent applications across over 220 patent families ranging from the core chemistry and materials of the nanopore itself to the manufacturing process of the Oxford Nanopore sequencers. This represents both broad and deep IP in nanopore sensing.
The technology performance results achieved by Oxford Nanopore in the past two years are ground-breaking and I believe the industry is only just starting to appreciate the magnitude of such achievements.
In November last year, Oxford Nanopore started shipping PromethION flow cells that included a number of technology upgrades, including work on the membrane to ensure higher levels of ‘pore occupancy’ (numbers of working channels in a flow cell). This month, a PromethION customer reported repeatedly achieving 220 Gb of sequencing data output per single Flowcell (the equivalent of a sequencing cartridge). At this yield, this equates to three human genomes per PromethION Flowcell at just under 30X coverage. As a single Flowcell can be purchased for as little as $625, this illustrates nanopore whole human genomes being accessible at a lower cost than traditional sequency-by-synthesis, but with richer data.
With a reminder that a PromethION device can run up to 48 of these flow cells at any time, this highlights the emerging ability to address the high-throughput end of the genomics market.
As these results become more common, this cost-competitive sequencing, delivering richer data than traditional short reads, promises a reshaping of the market.
By striking a strategic partnership with NVIDIA and integrating its high-end DGX AI server into its PromethION offering, Oxford Nanopore is ensuring that AI-powered genomics data analysis will provide a clear path towards continuous improvements in base-calling and support future developments in sequencing accuracy.
The company has also recently shown how customers using its most recent analysis algorithm, Bonito CRF, have achieved >98% single read accuracy; typically, single reads will be sequenced multiple times to generate high consensus accuracy. Furthermore, internal results from a new sequencing chemistry in development have already shown 99.1% single read accuracy. This chemistry is soon being released to early access users. At the same time SNP accuracy (single-nucleotide polymorphism is a substitution of a single nucleotide at a specific position in the genome present in at least 1% or more of the population) is now at 99.92% comparable to current short-read sequencing. For those of us who have been involved in this sector through our investment activities these achievements in long-read sequencing are staggering.
From early 2020, the company supported epidemiologists worldwide, with its MinION and GridION product lines of smaller sequencers, studying the SARS-CoV-2 evolution. As new variants of the coronavirus emerge, Oxford Nanopore will have a key role to play in COVID epidemiology – as it has done with other infectious diseases like Ebola and Yellow Fever and will do in the future for drug resistance across many pathogens.
During 2020, Oxford Nanopore started its journey into the regulated clinical markets by developing a new assay, LamPORE (https://nanoporetech.com/about-us/news/lampore-test-sars-cov-2-detection-gains-ce-ivd-mark) for COVID-19 testing – a test that showed ‘gold standard’ accuracy when used in more than 23,000 samples in the NHS. We’re excited that the company is now well set up with infrastructure to approach other applied markets.
Having moved in its new high-tech factory Oxford Nanopore is now able to satisfy the growing demand for its technology.

The technology performance improvements of the past few years coupled with the team’s great work in establishing the necessary infrastructure to build a global deep tech business will enable Oxford Nanopore to extend its market reach well beyond the current genomics research market and into multiple clinical and industrial high-volume applications. By achieving this I believe Oxford Nanopore has the potential to expand the sequencing market well beyond its current footprint and into the tens of $Bs, whilst taking a market leadership position in it.
The Long and the Short of it
Thanks to Oxford Nanopore’s technology performance achievements of the past few years long-read sequencing is now ready for prime time. We believe nanopore sequencing, with its ability to sequence long DNA fragments, in real time, at scale, represents the next generation of sequencing technologies and Oxford Nanopore has the potential to become its market leader. A market that is likely to grow well beyond what we can currently foresee.
